hv_approx#
- moocore.hv_approx(data, /, ref, *, maximise=False, nsamples=100000, seed=None, method='DZ2019-HW')[source]#
Approximate the hypervolume indicator.
Approximate the value of the hypervolume metric with respect to a given reference point assuming minimization of all objectives. The default
method="DZ2019-HW"is deterministic and ignores the parameterseed, whilemethod="DZ2019-MC"relies on Monte-Carlo sampling [1]. Both methods tend to get more accurate with higher values ofnsamples, but the increase in accuracy is not monotonic as shown in the example Comparing methods for approximating the hypervolume.See also
For details of the calculation, see the Notes section below.
- Parameters:
data (
ArrayLike) – Numpy array of numerical values, where each row gives the coordinates of a point. If the array is created from theread_datasets()function, remove the last column.ref (
ArrayLike) – Reference point as a 1D vector. Must be same length as a single point in thedata.maximise (
bool|Sequence[bool], default:False) – Whether the objectives must be maximised instead of minimised. Either a single boolean value that applies to all objectives or a list of booleans, with one value per objective. Also accepts a 1d numpy array with value 0/1 for each objectivensamples (
int, default:100000) – Number of samples for Monte-Carlo sampling. Higher values give more accurate approximation of the true hypervolume but require more time.seed (
int|Generator|None, default:None) – Either an integer to seednumpy.random.default_rng(), Numpy default random number generator (RNG) or an instance of a Numpy-compatible RNG.Noneuses the equivalent of a random seed (seenumpy.random.default_rng()).method (
Literal['DZ2019-HW','DZ2019-MC'], default:'DZ2019-HW') – Method to approximate the hypervolume.
- Returns:
float– A single numerical value, the approximate hypervolume indicator.
Notes
This function implements the methods proposed by Deng and Zhang[11] to approximate the hypervolume:
\[\widehat{HV}_r(A) = \frac{2\pi^\frac{m}{2}}{\Gamma(\frac{m}{2})}\frac{1}{m 2^m}\frac{1}{n}\sum_{i=1}^n \max_{y \in A} s(w^{(i)}, y)^m\]where \(m\) is the number of objectives, \(n\) is the number of weights \(w^{(i)}\) sampled, \(\Gamma()\) is the gamma function
math.gamma(), i.e., the analytical continuation of the factorial function, and \(s(w, y) = \min_{k=1}^m (r_k - y_k)/w_k\).In the default
method="DZ2019-HW", the weights \(w^{(i)}, i=1\ldots n\) are defined using a deterministic low-discrepancy sequence. The weight values depend on their number (nsamples), thus increasing the number of weights may not necessarily increase accuracy because the set of weights would be different. Inmethod="DZ2019-MC", the weights \(w^{(i)}, i=1\ldots n\) are sampled from the unit normal vector such that each weight \(w = \frac{|x|}{\|x\|_2}\) where each component of \(x\) is independently sampled from the standard normal distribution. The original source code in C++/MATLAB for both methods can be found here.References
Examples
>>> x = np.array([[5, 5], [4, 6], [2, 7], [7, 4]]) >>> moocore.hypervolume(x, ref=[10, 10]) 38.0 >>> moocore.hv_approx(x, ref=[10, 10], seed=42, method="DZ2019-MC") 38.01475 >>> moocore.hv_approx(x, ref=[10, 10], method="DZ2019-HW") 37.99989
Merge all the sets of a dataset by removing the set number column:
>>> x = moocore.get_dataset("input1.dat")[:, :-1]
Dominated points are ignored, so this:
>>> moocore.hv_approx(x, ref=10) 93.5533
gives the same hypervolume approximation as this:
>>> x = moocore.filter_dominated(x) >>> moocore.hv_approx(x, ref=10) 93.5533
The approximation is far from perfect for large number of dimensions:
>>> x = moocore.get_dataset("ran.10pts.9d.10")[:, :-1] >>> x = moocore.filter_dominated(-x, maximise=True) >>> x = moocore.normalise(x, to_range=[1, 2]) >>> reference = 0.9 >>> moocore.hypervolume(x, ref=reference, maximise=True) 0.483633123747 >>> moocore.hv_approx(x, ref=reference, maximise=True) 0.4852583123
Gallery examples#
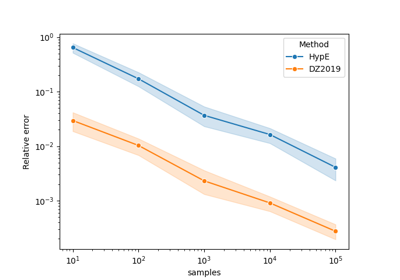
Comparing methods for approximating the hypervolume